Opinion 
 Creative Commons, CC-BY
Creative Commons, CC-BY
On The Importance of Open-Source Databases for NMRBased Metabolomics
*Corresponding author: Geerten W Vuister, Department of Molecular and Cell Biology, Leicester Institute of Structural and Chemical Biology,University of Leicester, United Kingdom.
Received: March 02, 2023; Published: March 23, 2023
DOI: 10.34297/AJBSR.2023.18.002455
Summary
We report on the examination of three public domain databases for NMR metabolomics. Our analysis shows several categories of problems across these databases. We are currently working on remediation and integration of the available data.
Keywords: Metabolomics, NMR, Database, Chemical Shift, Coupling, Deconvolution, Spin Systems
Abbreviations: NMR: Nuclear Magnetic Resonance; HMDB: Human Metabolome Data Base; BMRB: Biological Magnetic Resonance Data Bank: GISSMO; Guided Ideographic Spin System Model Optimization; SSMs: Spin System Matrices; 1D1H: 1 Dimensional Proton Spectra
Background
Metabolomics concerns the system wide study of all the metabolites, defined as small molecules with a molecular weight < 1.5KDa in a biological sample, with the aim to elucidate their role and significance in biological processes [1-3]. Metabolites can serve as important markers of both healthy and diseased cellular states, as well as characterise the metabolic fluxes. Mass spectrometry and Nuclear Magnetic Resonance (NMR) spectroscopy are the two premiere experimental techniques used in metabolomics studies. In NMR spectroscopy, a metabolite is identified by a characteristic signature of typically several peaks in the NMR spectrum, with each peak being described by its chemical shift, i.e. position in the spectrum expressed in ppm, its signal intensity and its width at half height expressed in Hz and its so-called coupling pattern. The spectra obtained in NMR-based metabolomics are comprised of the sum of all the signatures of the metabolites in the metabolic sample and typically consist of hundreds to even thousands of peaks. The resulting crowding and signal overlap renders the identification of the signals, i.e. the deconvolution of the experimental spectrum into the relative occurrence of the spectra of its constituent metabolites, a non-trivial exercise. Such a deconvolution is also crucially improved with the presence of reliable reference spectral data of all, or at least the majority, of metabolites. In order to discern biologically relevant information from the experimental metabolomics spectra of a biological sample, its peaks must be appropriately identified and annotated, i.e. assigned to the correct metabolite(s), often by referencing to the previously measured spectra of pure metabolite standards. Therefore, researchers aiming to perform NMR-based metabolomics must have access to such a collection of properly annotated metabolite spectra.
The two most well-known publicly accessible databases for NMR-based metabolomics are the Human Metabolome Data Base (HMDB) [4] which boasts > 200,000 metabolite entries with extensive biological metadata, but is primarily focused on Mass Spectrometry, and the Biological Magnetic Resonance Data Bank (BMRB) [5], an NMR-specific database containing > 2,700 experimentally supported entries. Commercial metabolomics databases are typically linked to NMR analysis software such as Chenomx [6] with annotated spectra for 336 compounds at a range of spectrometer frequencies and sample pHs or Know It All Metabolomics Edition [7] with spectra for ~160,000 compounds, many of which are pharmaceuticals. In general, these commercial databases tend to be more consistent and more properly annotated than public databases but require costly subscriptions, making them difficult to justify for academic use.
Given that a metabolite NMR spectrum will vary depending on the sample conditions, e.g. temperature, pH, solvent, and on the field strength of the spectrometer, the ideal database would need a reference for each metabolite for every value of every condition for all possible spectrometers. Of course, this would be practically infeasible, as it would be incredibly expensive and laborious. Alternatively, with good understanding of metabolite physio-chemical properties and NMR theory, using simulation one can potentially extrapolate from a single, completely annotated spectrum with its respective metabolite chemical data, to other conditions or other spectrometer field strengths. One such effort was made with the Guided Ideographic Spin System Model Optimization library (GISSMO) [8] which uses spin system matrices (SSMs) to generate a peak-list that represents an NMR spectrum and so represents the data independently of spectrometer field strength. In contrast, other databases typically store peak-lists measured from the experimental spectrum with limited peak to chemical structure annotation.
Methods
The CcpNmr Analysis Metabolomics program is part of a software suite for biomolecular NMR that also includes Analysis Assign [9] for general NMR data analysis and Analysis Screen [10] for NMR-based small molecule screening. As a part of our effort to develop Analysis Metabolomics, we examined the experimentally supported metabolite entries from the HMDB, BMRB and GISSMO. We harvested the available experimental 1D proton (1D1H) NMR spectra with associated annotation data, i.e. peak-lists from HMDB (n=717) and BMRB (n=1196) and SSM data from GISSMO (n=643), from their respective websites. Some manual remediation was required in this process, to alleviate data-retrieval issues. Just like the Analysis Assign and Analysis Screen programs, the Analysis Metabolomics program has inherent macro-writing and socalled “pipes” for automation of actions. In addition, the program installation contains a large number of relevant scientific python packages, including the nmrsim package [11] for spectral simulation. Using these tools, the peak-lists and SSM’s as obtained from the three databases, simulated spectra were generated for all entries and automatically compared against any available experimental spectra, accounting for moderate global misalignment and solvent/ reference artefact interference.
Results
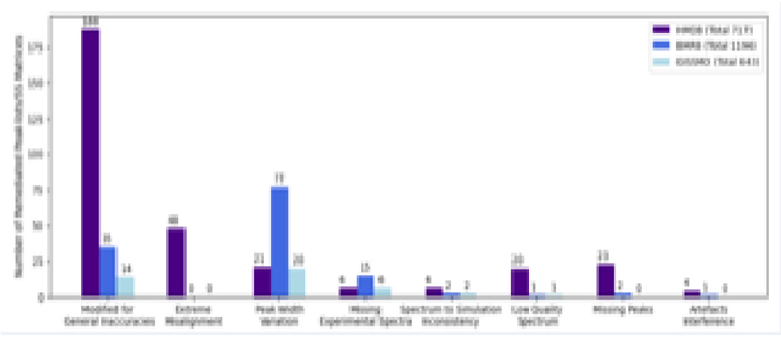
Our analysis revealed issues across all three databases (Figure 1). Unfortunately, we encountered missing or incorrect spectrum files in the database entries. Without an actual experimental spectrum, it is uncertain that the entry’s peak-list is accurate. For those entries with available experimental spectra, we visually inspected all those with a simulated vs experimental similarity score < 0.9. The most commonly observed problem was a too low precision in the peak chemical shift position, resulting in inaccurate simulated spectra. Other problems included improbable peakwidths, missing peaks, low-quality of the experimental data, and interference from experimental artefacts. As part of our efforts, we actively engage with the authors of these three databases to try to remedy the various errors. Overall, the number of actual available experimental 1D1H NMR spectra with sufficient annotation was also substantially lower than expected. The HMDB entries are overwhelmingly theoretical, with many entries containing no NMR spectra of any kind, while many of the BMRB entries did not contain peak height values for accurate recreation. One possible explanation is that NMR-based metabolomics spectra were never annotated with simulated recreation in mind, as public databases primarily use peak-lists for search purposes where such information is not vital to yield relevant hits. However, we feel that this dramatically limits database usefulness as there is little opportunity for users to interactively adapt the reference spectra to non-standard conditions, thus limiting confident identification to only the most well-known metabolites. Furthermore, peak-lists alone, even when recorded precisely, have limited simulation potential outside their immediate spectral and sample conditions, as extrapolation to other experimental conditions requires peak-to-atom assignment as a minimum.
Conclusion
With advancing open-source software tools for NMR-based metabolomics analysis and the general maturation of the field into (pre-)clinical practice, it is important that the publicly available databases that support these tools are robust and maintained with thorough and precisely annotated experimental spectra. Having identified the various issues, we are now building an integrated, remediated database of peak-list/SSM data, spectral metadata and metabolite metadata, using the three public databases as our primary source. This database will be open-source, freely available and hopefully further augmented through efforts of the community. Its features and release will be reported elsewhere in due course.
Acknowledgements
We thank the members of the CCPN development team, Ed Brooksbank, Luca Murredu, Vicky Higman, Eliza Ploskon and Gary Thomson (Univ. of Kent), for valuable discussion and feedback. We also thank Marie Phelan and Rudi Grossman (Univ. of Liverpool) for their continued support. MH is supported by a Leicester Institute of Structural and Chemical Biology PhD studentship. The research was supported by UKRI MRC grant MR/V000950/1 (to GWV).
Conflict of Interest
No conflict of interest.
References
- Rivera Velez SM, Navas J, Villarino NF (2021) Applying metabolomics to veterinary pharmacology and therapeutics. J Vet Pharmacol Ther 44(6): 855-869.
- Wishart DS (2019) Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol Rev 99(4): 1819-1875.
- Gonzalez Covarrubias V, Martinez Martinez E, del Bosque Plata L (2022) The Potential of Metabolomics in Biomedical Applications. Metabolites 12(2): 194.
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, et al. (2009) HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 37: D603-D610.
- Hoch JC, Baskaran K, Burr H, Chin J, Eghbalnia HR, et al. (2022) Biological Magnetic Resonance Data Bank. Nucleic Acids Res 51(D1): D368-D376.
- https://www.chenomx.com.
- https://sciencesolutions.wiley.com/software/.
- Dashti H, Westler WM, Tonelli M, Wedell JR, Markley JL, et al. (2017) Spin System Modeling of Nuclear Magnetic Resonance Spectra for Applications in Metabolomics and Small Molecule Screening. Anal Chem 89(22): 12201-12208.
- Skinner SP, Fogh RH, Boucher W, Ragan TJ, Mureddu LG, et al. (2016) CcpNmr AnalysisAssign: a flexible platform for integrated NMR analysis. J Biomol NMR 66(2): 111-124.
- Mureddu LG, Ragan TJ, Brooksbank EJ, Vuister GW (2020) CcpNmr AnalysisScreen, a new software programme with dedicated automated analysis tools for fragment-based drug discovery by NMR. J Biomol NMR 74(10-11): 565-577.
- https://pypi.org/project/nmrsim/.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.